negative sampling is an important step to identify negative training datasets that can be used for training and generate embeddings. In fact, it is such an important engineering practices that otherwise, it will be extremely computational difficult to calculate embeddings in simple techniques like randomwalk and others.
In the lecture 3.2-random walk approaches for node embeddings by Prof. Leskovec, he explained really well how random walk requires a normalization step that in theory requires V^2 computational complexity but can be approximated using negative sampling to k * V (in practice, k can ranges from 5 ~ 20).
The concept of negative sampling is not limited to graph, it is used in NLP – word2vec explained.
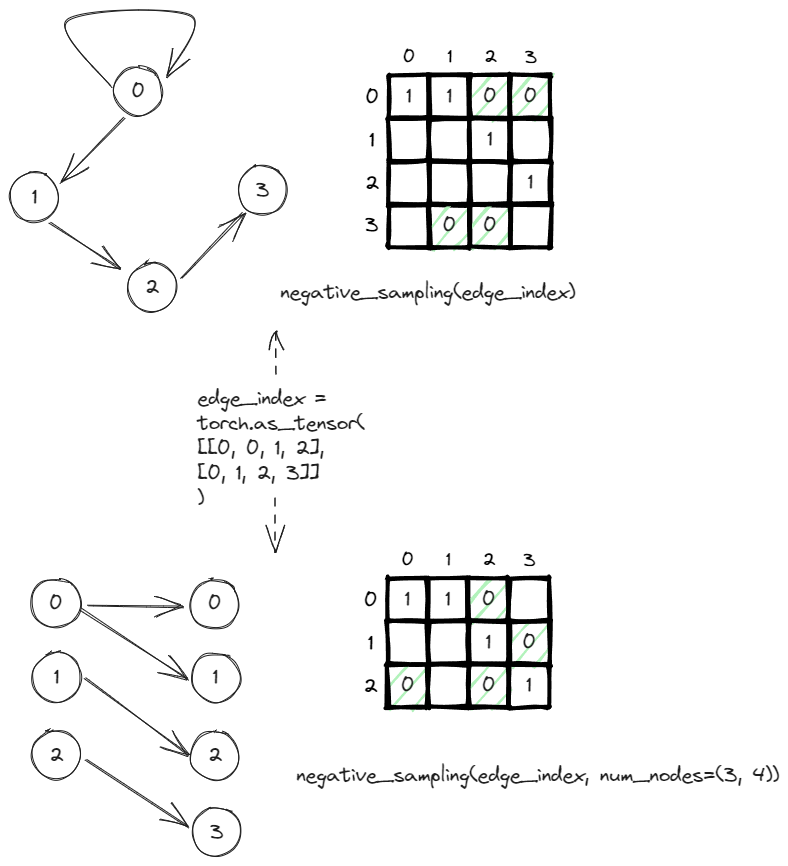
In order to do negative sampling in pyg, you can just call the negative_sampling function under torch_goemtric.utils. The only input it required is the edge_index which is a tensor containing the pairwise edges between from and to nodes.
In this example, the graph can be represented by the adjacency matrix. It is sparse and usually sparse because there exist only 4 edges out of 16 (V^2) possible edges. The rest of the nodes are implicitly negatives edges, edges that don’t and shouldn’t exist. The negative_sampling as default will return the same number of negative edges as the number of positives edges. In this case, it returns the four zeros (highlighted in green).
Also, if you pass a tuple (n,m) to the num_nodes, it indicates a graph is a bipartite graph where the number of sources nodes is n and the number of destination nodes is m. The adjacency matrix will be adjusted to n*m instead of n^2. The negatively sampling logic remain the same, it still returns 4 negatives edges that should not exist.
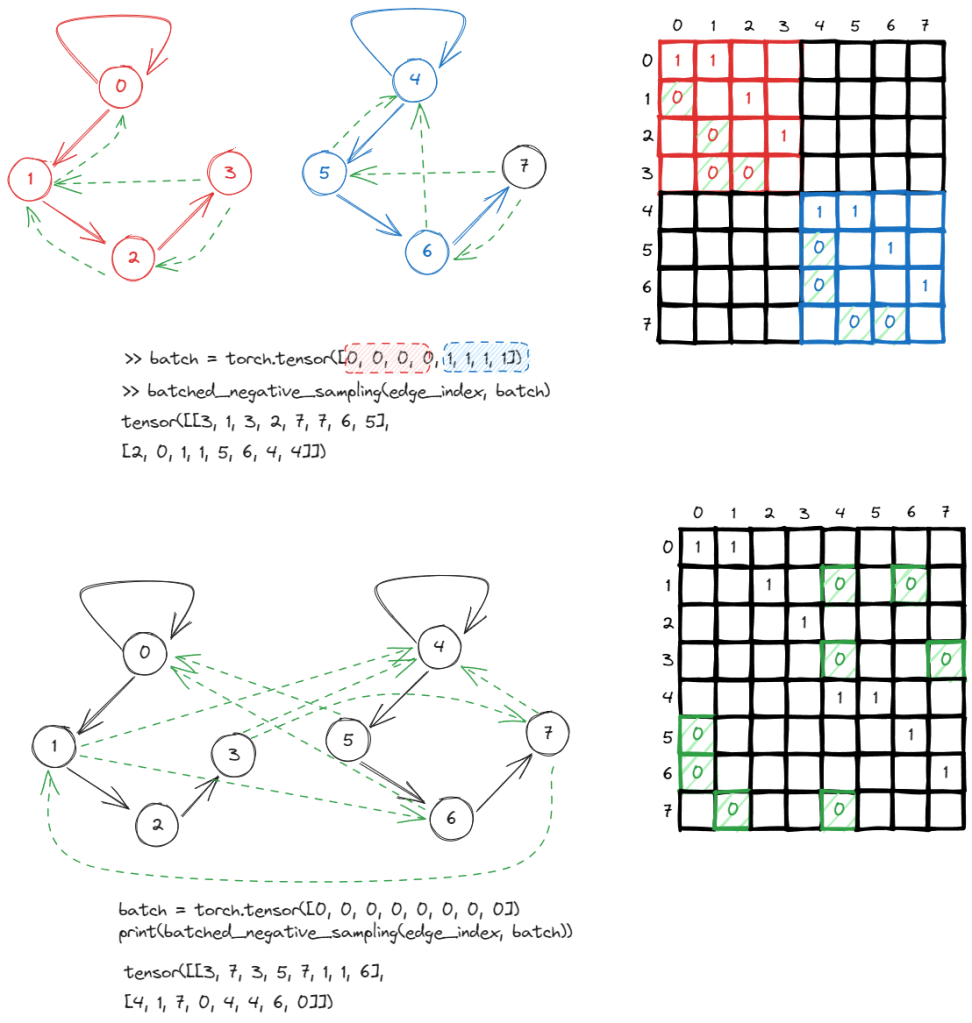
There is also a batch negative_sampling, the only difference is that you can pass in a batch parameter in order to distinguish which graph certain edges belong to. The negative sampling will only occur within the same graph instead of cross graphs.
To demonstrate, to treat the two graphs as one graph, you can simply ignore the the batch parameter and immediately, one start observing negative samples crossing graphs.
I found the chatgpt’s explanation easier to understand.
“
In your example, the batch tensor is torch.tensor([0, 0, 0, 0, 1, 1, 1, 1]). It means you have a batch of two graphs. The first four nodes (indices 0, 1, 2, 3) belong to the first graph (batch 0), and the second four nodes (indices 4, 5, 6, 7) belong to the second graph (batch 1).
So, when you’re using batched_negative_sampling(edge_index, batch), this function will generate negative edges (non-existing edges used for training graph learning models) while making sure the nodes of these negative edges are within the same graph. In other words, it will not generate a negative edge between nodes from different graphs.
“